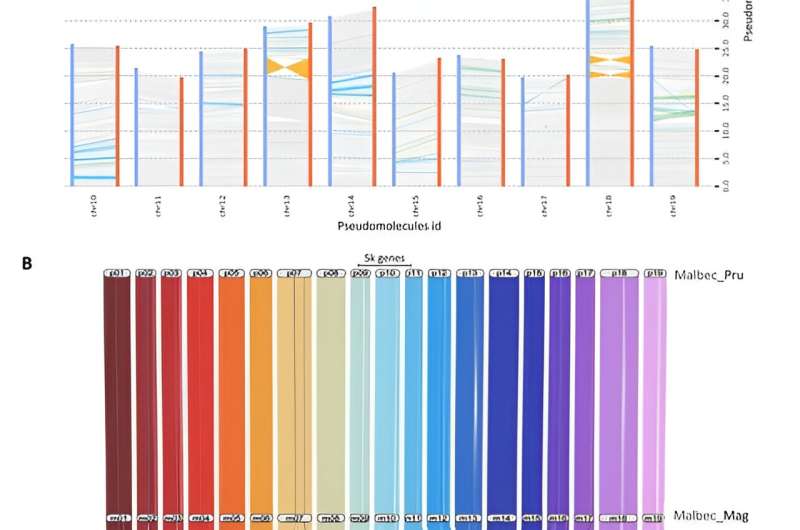
Synteny analysis based on comparison of 19 clustered pseudomolecules for Malbec haplophases. Credit: Horticulture Research (2024). DOI: 10.1093/hr/uhae080
Grapevine genomes, known for their high heterozygosity, present significant challenges for accurate assembly. Traditional approaches often focus on nearly homozygous lines, which fail to capture the full genetic diversity of complex cultivars such as Malbec.
Understanding clonal phenotypic variation, which significantly affects wine quality, adds to this complexity. To address these challenges, a detailed and accurate genomic assembly is essential to understand the genetic mechanisms that drive clonal variation and contribute to the unique attributes of the Malbec grape vine.
A team of researchers from the Instituto de Biología Agrícola de Mendoza, the Instituto de Ciencias de la Vid y del Vino and the Max Planck Institute for Biology Tübingen, among others, have published a study in the journal. Horticulture Research on March 14, 2024. This study presents the first diploid genome assembly of the Malbec grape vine, utilizing advanced sequencing technologies to resolve the two haplotypes inherited from its parental cultivars.
The study achieved a high-quality genome assembly of the Malbec grape vine using advanced PacBio long-read sequencing and triplication techniques. This method enabled the separation and assembly of two haplotypes inherited from the Malbec parent cultivars, Prunelard and Magdeleine Noire des Charentes.
The assembled genome revealed significant polymorphic regions and provided detailed gene pattern annotations for both haplotypes. Transcriptomic analysis of Malbec clonal variants revealed differences in gene expression, particularly highlighting higher anthocyanin content in certain clones. This increased anthocyanin content was associated with increased abscisic acid responses, leading to the overexpression of genes involved in phenylpropanoid metabolism and abiotic stress responses.
These findings underscore the critical role of haplotype-resolved assemblies in understanding the genetic basis of clonal variation and its impact on traits essential for wine quality and grapevine fitness.
Dr. Luciano Calderón, one of the lead researchers, said: “This genome assembly not only increases our understanding of the genetic diversity of Malbec, but also provides a valuable resource for studying the molecular mechanisms underlying clonal variation. It opens up possibilities new for breeding and improving vine cultivars”.
The assembled Malbec genome provides a comprehensive reference for future genetic studies and breeding programs aimed at improving grape cultivars. By understanding the genetic basis of traits such as fruit composition and stress responses, researchers can develop more resilient and high-quality vines, ultimately benefiting the wine industry and addressing the challenges posed by climate change.
More information:
Luciano Calderón et al, Diploid Genome Assembly of Malbec Grapevine Cultivar Enables Haplotype-Aware Analysis of Transcriptomic Differences Underlying Clonal Phenotypic Variation, Horticulture Research (2024). DOI: 10.1093/hr/uhae080
Provided by the Chinese Academy of Sciences
citation: From Vine to Wine: Decoding Malbec Genetic Diversity for Clonal Excellence (2024, June 28) Retrieved June 28, 2024 from https://phys.org/news/2024-06-vine-wine-decoding-malbec-genetic .html
This document is subject to copyright. Except for any fair agreement for study or private research purposes, no part may be reproduced without written permission. The content is provided for informational purposes only.
#Vine #Wine #Decoding #Malbecs #Genetic #Diversity #Clonal #Excellence
Image Source : phys.org
